性别: 男
职称: 教授
电子邮件: [email protected]
导师类型: 博士生导师
学科方向:071001-植物学
获得国家级高层次人才青年项目、上海市浦江人才等多个人才项目资助。主持多项国家自然科学基金委、上海市科委和上海市教委项目,获得省部级自然科学一等奖(排第3),并参与国家级一流本科课程教学。在科研方面,长期以来聚焦于从多维度基因组可塑性角度解析适应性表型性状和物种多样性形成的进化机制与分子基础,已发表SCI论文60余篇,主要代表成果发表于Nature Genetics, Nature Communications, Molecular Plant, New Phytologist, Plant Communications和Plant Journal等期刊。具体研究内容包括以下三个方面:
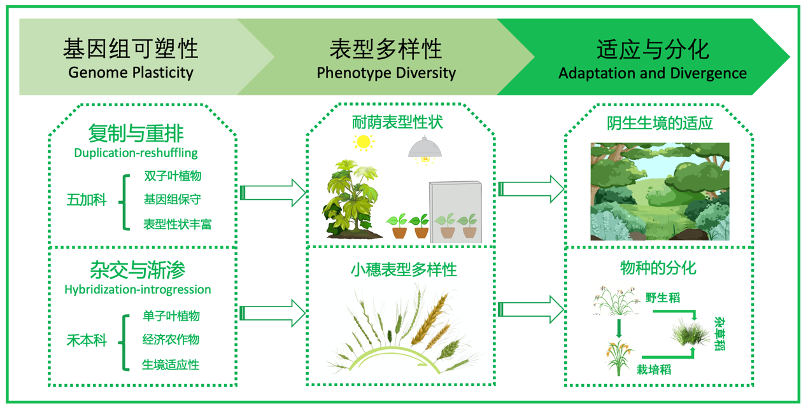
(1) 以双子叶五加科和单子叶禾本科为研究对象,展开大规模比较基因组学分析,揭示“复制与重排”和“杂交与渐渗”等进化机制,促进现存物种基因组在DNA线性水平可塑性的形成;
(2) 从全基因组水平开展比较表观遗传学分析,解析祖先基因组的染色质拓扑结构重塑、基因表达差异与甲基化修饰等表观遗传调控,增加现存物种基因组多维度可塑性形成的机制;
(3) 聚焦于植物防御、耐荫表型与杂草性状等关键表型,揭示“复制与重排”和“杂交与渐渗”等进化机制,促进关键适应性表型性状形成的分子基础 (详见图示)。
代表性论文(第一/通讯)
Li, L.F.#, Pusadee, T#, Wedger, M.J., Li, Y.L., Li, M.R., Lau, Y.L., Yap, S.J., Jamjod, S., Rerkasem, B., Hao, H., Song, B.K., Olsen, K.M. (2024) Porous borders at the wild-crop interface promote weed adaptation in Southeast Asia. Nature Communications 15, 1182
Hao, Y., Wang, X.F., Guo, Y.L., Li, T.Y., Yang, J., Ainouche, M.L., Salmon, A., Ju, R.T., Wu, J.H.*, Li, L.F.*, and Li, B.* (2024) Genomic and phenotypic signatures provide insights into the wide adaptation of a global plant invader. Plant Communications 5, 100820
Wang, X.F., Zhang, Y.X., Niu, Y.Q., Sha, Y., Wang, Z.H., Zhang, Z.B., Yang, J., Liu, B.* and Li, L.F.*. (2023) Post‐hybridization introgression and natural selection promoted genomic divergence of Aegilops speltoides and the four S‐genome diploid species. The Plant Journal 115, 1500-1513
Wang, Z.H., Wang, X.F., Lu, T.Y., Li, M.R., Jiang, P., Zhao, J., Liu, S.T., Fu, X.Q., Wendel, J.F., Van de Peer*, Liu, B*, and Li, L.F.*. (2022) Reshuffling of the ancestral core-eudicot genome and its role in shaping epigenetic modification in extant Panax. Nature Communications 13, 1902.
Li, L.F.*,#, Zhang, Z.B#, Wang, Z.H., Li, N., Sha, Y., Wang, X.F., Ding, N., Li, Y., Zhao, J., Wu, Y., Gong, L., Mafessoni, F., Levy, A.A.*, and Liu, B*. (2022) Genome sequences of the five Sitopsis species of Aegilops and the origin of polyploid wheat B-subgenome. Molecular Plant 15(3): 1-16.
Li, M.R., Ding, N., Lu, T., Zhao, J., Wang, Z.H., Jiang, P., Liu, S.T., Wang, X.F., Liu, B.*, and Li, L.F.* (2021) Evolutionary contribution of duplicated genes to genome evolution in the ginseng species complex. Genome Biology and Evolution 13: evab051.
Li, L.F., Cushman, S.A., He, Y.X., and Li, Y.* (2020) Genome sequencing and population genomics modeling provide insights into the local adaptation of weeping forsythia. Horticulture Research 7:130.
Li, M.R., Wang, H.Y., Ding, N., Lu, T., Huang, Y.C., Xiao, H.X., Liu, B. and Li, L.F.*. (2019) Rapid diverence followed by adaptation to contrasting ecological niches of two closely related columbine species Aquilegia japonica and A. oxysepala. Genome Biology and Evolution 11:919-930.
Li, L.F., Li Y.L., Jia Y.L., Caicedo A.L.*, and Olsen K.M.* (2017) Signatures of adaptation in the weedy rice genome. Nature Genetics 49: 811-814.
Li, M.R., Shi, F.X., Li, Y.L., Jiang, P., Jiao, L.L., Liu, B.*, and Li, L.F.*. (2017). Genome-wide variation patterns uncover the origin and selection in cultivated ginseng (Panax ginseng Meyer). Genome Biology and Evolution 9:2159-2169.
Wang, X., Zhang, H., Li, Y., Zhang, Z., Li, L.F.*, and Liu, B*. (2016). Transcriptome asymmetry in synthetic and natural allotetraploid wheats, revealed by RNA-sequencing. New Phytologist 209: 1264-1277.
Li, M.R., Shi, F.X., Zhou, Y.X., Li, Y.L., Wang, X.F., Zhang, C., Wang, X.T., Liu, B., Xiao, H.X.*, and Li, L.F.* (2015). Genetic and epigenetic diversities shed light on domestication of cultivated ginseng (Panax ginseng CA Meyer). Molecular Plant. 8: 1612-1622.
Li, L.F.*, Liu, B., Olsen, K.M., and Wendel, J.F. (2015). A re-evaluation of the homoploid hybrid origin of Aegilops tauschii, the donor of the wheat D-subgenome. New Phytologist 208:4-8.
Li, L.F.*,#, Wang, H.Y.#, Pang, D., Liu, Y., Liu, B., and Xiao, H.X.* (2014). Phenotypic and genetic evidence for ecological speciation of Aquilegia japonica and A. oxysepala. New Phytologist 204:1028-1040.



